Datasets |
Code for the masses |
|
The validation of newly designed methods and protocols relies on the ability to apply them to known datasets. While one or even several datasets cannot account for all instances, knowing how new ideas compare to previous results is a first step. Some of the provided datasets are well known while others are from research conducted at exeResearch LLC. The datasets are available from the exeResearch GitHub site.
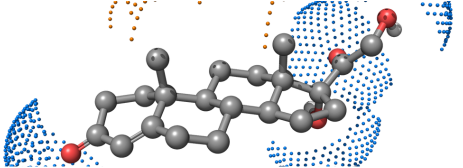
CoMFA Steroid Benchmark Dataset
This steroid dataset was made famous by the original CoMFA article (J Am Chem Soc, 1988, 110(18), pp5959-5967. DOI:10.1021/ja00226a005). The provided steroid dataset is the corrected version by Eugene A Coats (Perspect Drug Discovery Des, 1998, 12/13/14, pp199-213. DOI:10.1023/A:1017050508855). Available here! 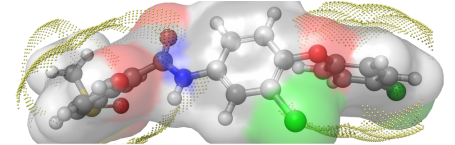
Selwood Dataset The Selwood dataset is well known to those interested in genetic algorithms and QSAR modeling. This is the Selwood dataset used in the Rogers and Hopfinger Genetic Function Approximation (GFA) study (J Chem Inf Comput Sci, 1994, 34(4), pp854-866. DOI:10.1021/ci00020a020); originally curated by Selwood et al. (J Med Chem, 1990, 33(1) pp136-142. DOI:10.1021/jm00163a023). Available here! 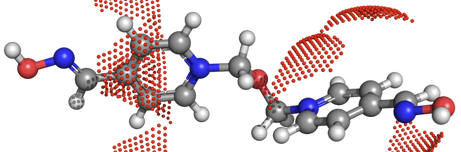
Oxime Dataset
The oxime dataset contains 17 oximes with percent reactivation values for cyclosarin, sarin, tabun, and VX. The conformation and AM1-bcc atomic charges are provided as a stacked MOL2 file and the conformations and the percent reactivation values are provided in a SDFile. This dataset is from Esposito et al. Chem Res Tox, 2014, 27(1), pp99-110. DOI:10.1021/tx400350b Available here! |
The following functions/applications are commonly used in our research projects. A brief description of each function is provided below. Complete instructions are provided within the individual files along with full instructions within the file. The following code is available from the exeResearch GitHub site.
MACCS Key Counts (SVL)
Counts the occurrence of the 166 MACCS keys for each molecule in a MOE database (mdb). The number of each key's occurrence is written to a user specified CSV file. The option to include the compounds' names and endpoints in the CSV file is available from the opts variable. Download Set Amide Bonds to 180 Degrees (SVL)
It is common for small drug-like compounds and peptides that are constructed from SMILES strings, and even SDfiles, to have amide bonds where the hydrogen of the nitrogen atom and the oxygen of the carboxyl group result in a dihedral angle of approximately 0 degrees. This SVL function sets all amide dihedral angles to 180 degrees and provides the option for energy minimization of the new conformation. The interactive (MOE 3D window) version allows the user to select the amide bond of interest while the mdb version of the function indicates, in a new column (field) labeled "numDihedChanged," the number of changed amide bonds. Download |
